Abstract
AML is a heterogeneous clonal disorder that is characterized by the accumulation of complex genomic abnormalities that impact disease biology and outcome. Despite significant advances in our understanding of the impact of these mutations on survival (OS), routine AML risk stratification is based primarily on cytogenetic analyses and a limited number of genes. This approach does not take into account the complexity and the interaction between these mutations and how particular constellations of genomic and clinical risk factors affect a patient's (pt) outcome.
We developed a novel prognostic model that incorporates clinical, cytogenetic, and molecular mutational data to determine personalized (precision) outcomes specific to a particular pt.
Genomic and clinical data were obtained from a publically available dataset (GitHub - gerstung-lab/AML-multistage, Gerstung M et al, Nature Genetics 2017) that contains genomic sequencing data on 111 genes along with clinical and treatment data from 1,540 pts with AML enrolled in 3 different trails. The developed model was then applied to a validation cohort of 468 pts (283 pts treated at our institution and 185 pts from the TCGA Atlas). A univariate cox regression analysis was used to evaluate the impact of each mutation on OS. A random survival forest (RSF) algorithm was used to build the new model, in which clinical and molecular variables were randomly selected for inclusion in determining survival. Survival prediction was thus specific to each pt's particular clinical and molecular characteristics. C-index was used to evaluate the accuracy of the new model compared to ELN risk classification.
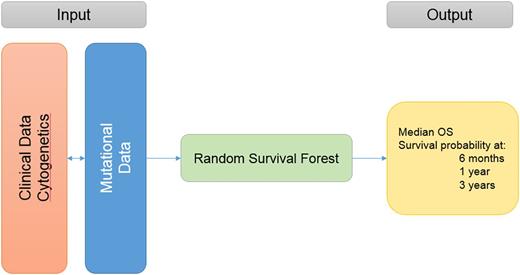
A total of 1413 pts with complete cytogenetic analysis were included in the training cohort. The median age was 57 years (range, 18-65); 168 pts (12%) had favorable risk cytogenetics per ELN criteria, 1012 (72%) intermediate (INT), and 233 (16%) adverse. The most commonly mutated genes were: NPM1 (28%), DNMT3A (24%), FLT3-ITD (21%), NRAS (18%), FLT3-TKD (13%), TET2 (12%), and IDH2 (10%). Mutations occurred in different frequencies and had different impact on OS in each risk group. The most commonly mutated genes in the favorable risk group were: NRAS (32%), KIT (23%), FLT3-TKD (20%), FLT3-ITD (17%), WT1 (10%), and TET2 (7%). Only mutations in KIT (HR 1.9, p=.02), TET2 (HR 2.4, p=.02), and DNMT3A (HR 3.2, p=.04, occurring in only 2% of pts) had a negative impact on OS in this group. The most commonly mutated genes in the INT risk group were: NPM1 (38%), DNMT3A (30%), FLT3-ITD (24%), NRAS (15%), TET2 (15%), FLT3-TKD (14%), IDH2 (13%) and RUNX1 (11%). Mutations in NPM1 (HR .84, p=.05), FLT3-ITD (HR 1.8, p <.001 ), FLT3-TKD (HR .66, p=.002), CEBPA (HR .5, p <.001), TET2 (HR 1.3, p=.03), RUNX1 (HR 1.5, p=.02), IDH1 (HR 1.4, p=.04 ), SRSF2 (HR 1.9, P <.001), STAG2 (HR 1.5, p=.02), ASXL1 (HR 1.8, p <.001) and RAD21 (HR .5, p <.01) impacted OS. The most commonly mutated genes in pts with adverse cytogenetics included: TP53 (30%), NRAS (20%), DNMT3A (12%), PTPN11 (12%), RUNX1 (11%), FLT3-ITD (10%), and KRAS (8%). Only mutations in TP53 (HR 2.1, p <.001) and TET2 (HR .4, p=.03) impacted OS. All genomic-clinical variables were included in the RSF algorithm. Variable importance analyses (the most important variables that contributed to the outcome) and multiple backward elimination analyses (identifying the least number of variables that can provide the least error rate) identified the following variables that impacted OS: cytogenetics, transplant (yes vs. no), hemoglobin, WBC, bone marrow blast %, peripheral blast %, performance status, ASXL1, FLT3-ITD, FLT3-TKD, CEBPA, DNMT3A, TET2, RUNX1, TP53, NPM1, IDH1, IDH2, PHF6, SRSF2, ZRSR2, and U2AF1 . The clinical and mutational variables can be entered into a web application that can run the trained model and provide OS estimates as an output, Figure 1.The C-index for the new model was 0.71 which outperformed ELN classification (C-index 0.64). When applying the new model to the validation cohort, the C-index was 0.70.
Genomic alterations have a differential impact on OS in each cytogenetic risk group, highlighting the complexity of incorporating these mutations into risk stratification. A personalized prediction model based on clinical-genomic data can accurately provide survival unique to each individual pt and can outperform ELN classifications. The web application that can ease the translation of this model into the clinic is being developed.
Advani: Takeda/ Millenium: Research Funding; Pfizer: Consultancy. Gerds: Incyte: Consultancy; CTI BioPharma: Consultancy. Sekeres: Celgene: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal